Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction (PCR)
Definition:
PCR is an enzymatic process in which a specific region of DNA is replicated over and over again to yield many copies of a particular sequence. This method combines the principles of complementary nucleic acid hybridization with those of nucleic acid replication applied repeatedly through numerous cycles. The purpose of a PCR (Polymerase Chain Reaction) is to make a huge number of copies of a gene.
Two methods currently exist for amplifying the DNA or making copies
- Cloning—takes a long time for enough clones to reach maturity
- PCR—works on even a single molecule quickly
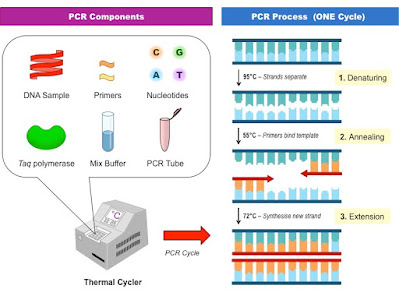 |
| PCR process |
History And Discovery:
PCR was invented by Kary Mullis. At the time he thought up PCR in 1983, Mullis was working in Emeryville, California for Cetus, one of the first biotechnology companies. There, he was charged with making short chains of DNA for other scientists.
Mullis has written that he conceived of PCR while cruising along the Pacific Coast Highway 128 one night on his motorcycle. He was playing in his mind with a new way of analyzing changes (mutations) in DNA when he realized that he had instead invented a method of amplifying any DNA region. Mullis has said that before his motorcycle trip was over, he was already savoring the prospects of a Nobel Prize. He shared the Nobel Prize in chemistry with Michael Smith in 1993.
Principle of PCR:
The target sequence of nucleic acid is denatured to single strands, primers specific for each target strand sequence are added, and DNA polymerase catalyzes the addition of deoxynucleotides to extend and produce new strands complementary to each of the target sequence strands (cycle 1).
In cycle 2, both double-stranded products of cycle 1 are denatured and subsequently serve as targets for more primer annealing and extension by DNA polymerase.
 |
| PCR cycle |
Requirements for PCR:
- A PCR reaction contains the target double-stranded DNA
- Two primers that hybridize to flanking sequences on opposing strands of the target
- All four deoxyribonucleoside triphosphates
- Buffer, co-factors of enzyme and water
- DNA polymerase ( Heat-stable Taq polymerase from the thermophilic bacterium Thermus aquaticus)
Steps Involved in PCR:
A. Extraction and Denaturation of Target Nucleic Acid
For PCR, the nucleic acid is first extracted (released) from the organism or a clinical sample potentially containing the target organism by heat, chemical, or enzymatic methods.
Once extracted, target nucleic acid is added to the reaction mix containing all the necessary components for PCR (primers, nucleotides, covalent ions, buffer, and enzyme) and placed into a thermal cycler to undergo amplification.
B. Steps in Amplification
1.Denaturation
The reaction mixture is heated to 95°C for a short time period (about 15–30 sec) to denature the target DNA into single strands that can act as templates for DNA synthesis.
2.Primer annealing
This annealing temperature is calculated carefully to ensure that the primers bind only to the desired DNA sequences (usually around 55 degrees C).
Primers are short, single-stranded sequences of nucleic acid (i.e., oligonucleotides usually 20 to 30 nucleotides long) selected to specifically hybridize (anneal) to a particular nucleic acid target, essentially functioning like probes.
The mixture is rapidly cooled to a defined temperature which allows the two primers(Forward and Backward) to bind to the sequences on each of the two strands flanking the target DNA.
One primer binds to each strand. The two parental strands do not re-anneal with each other because the primers are in large excess over parental DNA.
3.Extension
The temperature of the mixture is raised to 72°C (usually) and kept at this temperature for a pre-set period of time to allow DNA polymerase to elongate each primer by copying the single-stranded templates.
Taq polymerase is the enzyme commonly used for primer extension, which occurs at 72°C. This enzyme is used because of its ability to function efficiently at elevated temperatures and to withstand the denaturing temperature of 94°C through several cycles.
Annealing of primers to target sequences provides the necessary template format that allows the DNA polymerase to add nucleotides to the 3’ terminus (end) of each primer and extend sequence complementary to the target template.
The three steps of the PCR cycle are repeated. Thus in the second cycle, the four strands denature, bind primers, and are extended. No other reactants need to be added. The three steps are repeated for a third cycle and so on for a set of additional cycles.
C. Product Analysis
Gel electrophoresis of the amplified product is commonly employed after amplification. The amplified DNA is electrophoretically migrated according to their molecular size by performing agarose gel electrophoresis. The amplified DNA forms clear bands which can be visualized under ultra-violet (UV) light.
Youtube Video -
Applications of PCR:
Types of PCR (Polymerase chain reaction):
1.Nested PCR
Principle: Nested PCR is a useful modification of PCR technology where the specificity of the reaction is enhanced by preventing the non-specific binding with the help of the two sets of primers.
The first set of primers binds outside of our target DNA and amplifies larger fragments while another set of primers binds specifically at the target site. In the second round of amplification, the second set of primers amplifies only the target DNA.
Application: Nested PCR is a helpful method for phylogenetic studies and the detection of different pathogens. The technique has higher sensitivity; hence even if the sample contains lower DNA, it can be amplified which is not feasible in the conventional PCR technique.
2.Real-Time PCR (Quantitative PCR (qPCR)
Principle: Quantitative PCR (qPCR), also called real-time PCR or quantitative real-time PCR, is a PCR-based technique that couples amplification of a target DNA sequence with quantification of the concentration of that DNA species in the reaction.
Conventional PCR is a time-consuming process where the PCR products are analyzed through gel electrophoresis. qPCR facilitates the analysis by providing real-time detection of products during the exponential phase.
The principle of real-time PCR depends on the use of fluorescent dye. The concentration of the nucleic acid present in the sample is quantified using the fluorescent dye or using the fluorescent-labeled oligonucleotides.
Application: Q-PCR is applied in genotyping and quantification of pathogens, microRNA analysis, cancer detection, microbial load testing, and GMOs detection.
3.Reverse Transcriptase PCR (RT-PCR)
Principle: Reverse transcription PCR (RT-PCR) is a modification of conventional PCR, whereby RNA molecules are first converted into complementary DNA (cDNA) molecules that can then be amplified by PCR.
In RT-PCR, the RNA template is first converted into complementary DNA (cDNA) using reverse transcriptase. The cDNA then acts as a template for exponential amplification using PCR.
RT-PCR can be conducted either in a single tube or as two steps in different tubes. The one-step method is more effective with fewer chances of contamination and incorporation of variations.
Application: RT-PCR is used in research methods, gene insertion, genetic disease diagnosis and cancer detection.
4.Inverse PCR
Principle: Inverse polymerase chain reaction (Inverse PCR) is one of the many variants of the polymerase chain reaction that is used to amplify DNA when only one sequence is known.
Conventional PCR requires primers complementary to both terminals of the target DNA, but Inverse PCR allows amplification to be carried out even if only one sequence is available from which primers may be designed.
The inverse PCR involves a series of restriction digestion followed by ligation, which results in a looped fragment that can then be primed for PCR through a single section of known sequence. Then, like other polymerase chain reaction processes, the DNA is amplified by the temperature-sensitive DNA polymerase.
Application: Inverse PCR is especially useful for the determination of insert locations of various transposons and retroviruses in the host DNA.
5.Asymmetric PCR
Principle: Asymmetric PCR is a variation of PCR used to preferentially amplify one strand of the original DNA more than the other.
Asymmetric PCR differs from regular PCR by the excessive amount of primers for a chosen strand. As the asymmetric PCR progresses, the lower concentration limiting primer is quantitatively incorporated into newly synthesized double-stranded DNA and used up. Consequently, linear synthesis of the targeted single DNA strand from the excess primer is formed after depletion of the limiting primer.
Application: It is useful when amplification of only one of the two complementary strands is needed, such as in sequencing and hybridization probing.
References:
1.Sastry A.S. & Bhat S.K. (2016). Essentials of Medical Microbiology. New Delhi: Jaypee Brothers Medical Publishers.
2.David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
3.Bailey, W. R., Scott, E. G., Finegold, S. M., & Baron, E. J. (1986). Bailey and Scott’s Diagnostic microbiology. St. Louis: Mosby.
Bottom Line:
All pieces of information of this post are collected from reliable sources. You can use this blog post with a proper citation! If you want to add more information here, comment below now.



No comments